Projects
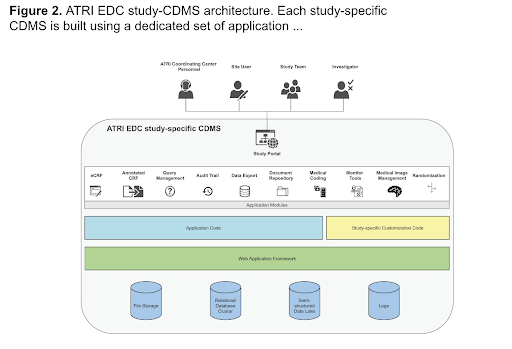
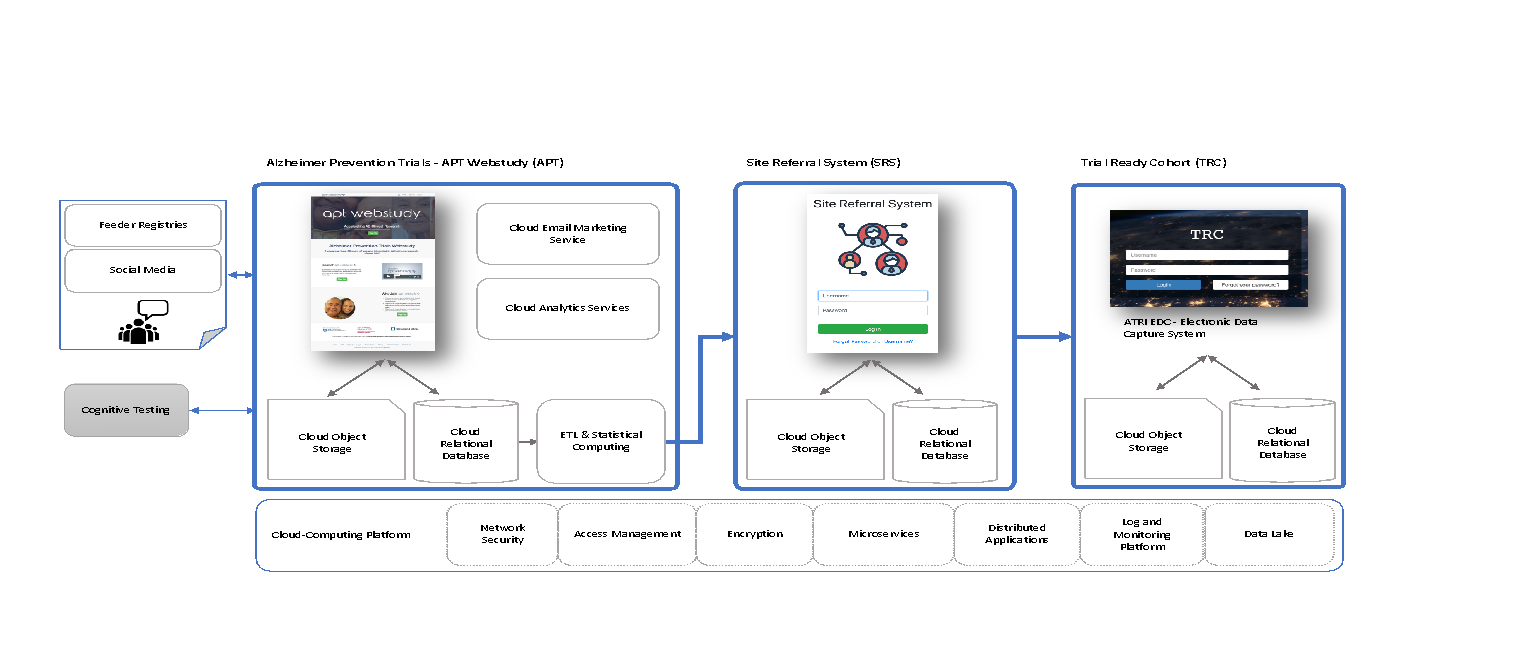
ATRI Electronic Data Capture System (ATRI EDC)
The ATRI EDC system was developed, tested, and validated using community-based agile software development methods and cloud-native single-page application design principles. It offers an increasing number of application modules, supports a high degree of study-specific configuration, and empowers study teams to effectively communicate and collaborate on the accurate and timely completion of study activities.
The ATRI EDC system has several advantages: its modular capabilities can accommodate rapidly evolving research designs and technologies; its community-based agile development approach and community-friendly licensing model encourage collaboration per the principles of open science; finally, with continued development and community building efforts, the system has the potential to facilitate the effective conduct of clinical studies beyond the field of AD/ADRD.
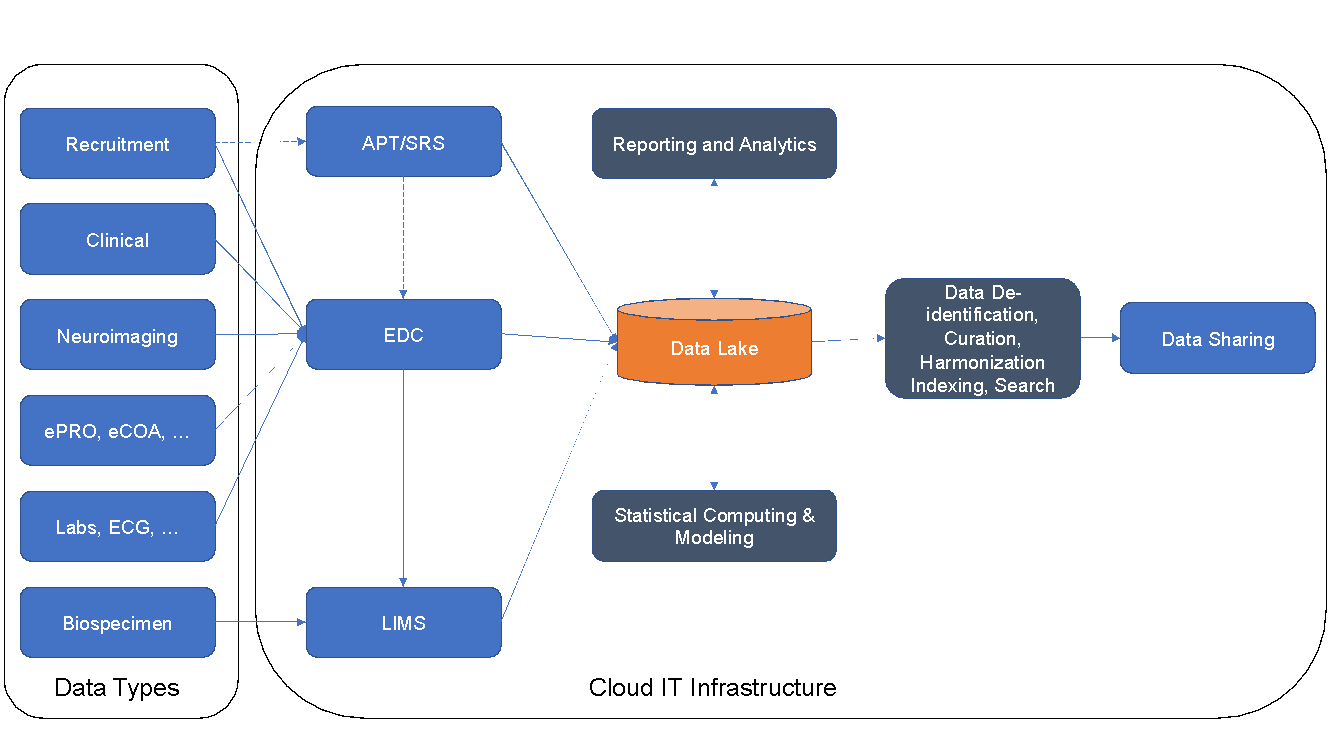
Core Modules: eCRFs, Annotated CRFs, Query Management, Audit Trail, Document Management, Data Export, Standard Reporting, and Data Lake
Extensions: Clinical monitor visit reporting, Adverse Event coding, Image management, and Safety reporting
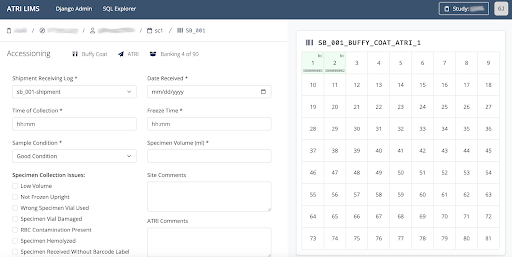
ATRI Laboratory Information Management System (ATRI LIMS)
Launched in 2023, the Laboratory Information Management System (LIMS) is a product of a collaboration between the ATRI Informatics and Biomarker teams to manage specimen data and inventory. ATRI LIMS offers a new, robust way of keeping track of specimen samples while recording pertinent data for fast information recall and preserving the specimen chain of custody.
Biospecimen management
- Full life-cycle
- Cloud-native, Secure, Scalable
- Audit trails
Part of lab automation initiative
- Efficient workflows
- Robotics, IOT, Instruments
Integration with clinical trials IT systems
- Coordinated data flows via APIs
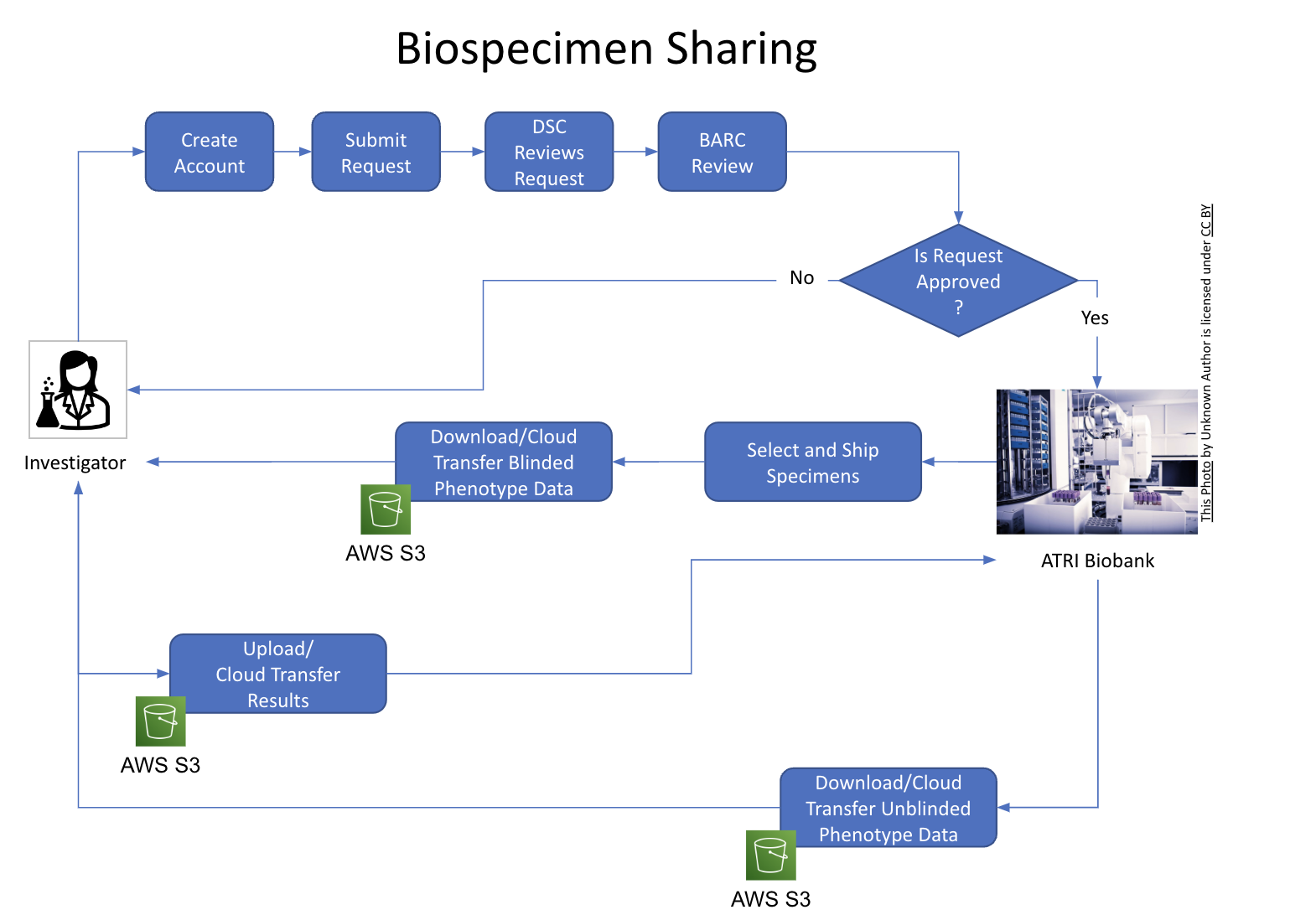
ATRI Data Sharing Platform
Goal: Accelerate clinical trials neuroimaging life-cycle from acquisition to data sharing
Challenges:
- Data sets are large and heterogeneous
- Advanced de-identification processes are required to protect participant privacy and confidentiality
- Analysis requires powerful computational resources, significant expertise, and specialized tools\
Collaboration with HMS/MGH (Sperling, Schultz) and ADIR-Mayo Clinic (Jack, Schwartz):
- Site certification and acquisition
- Data quality
- De-identification and defacing
- Normalization and packaging
- Reformatting (NIfTI)
- Reconciliation of results with clinical dataset
- Integration of analysis results and derived data
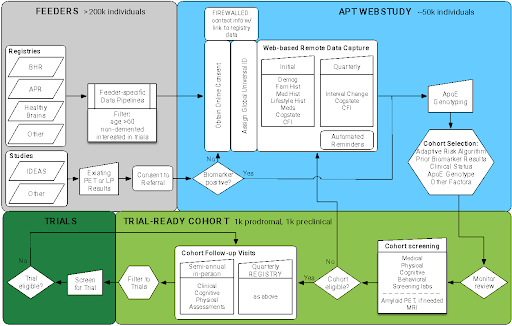
APT Webstudy
Launched end of December 2017
Longitudinal remote participant lifestyle and cognitive assessments
Features:
- HIPAA-compliant
- Web-based participant consent and assessments
- Mobile-first
- Social login
- User engagement
- Multi-language support
Recruitment efforts:
- Brain Health Registry (BHR)
- Alzheimer’s Prevention Registry (APR)
- HealthyBrains.org
- TrialMatch
- Media (digital/local)
- Community Outreach
- Site referrals
Retention:
- Low barriers (10-15 min per visit, flexible schedule)
- Engagement: feedback on progress, testing
- Education: updates on trials
ATRI Informatics Platform
The ATRI-IP serves as an extensible, secure, and scalable information, collaboration, and process management infrastructure to facilitate the effective execution of the program- and study-related research activities by investigators, study teams, and performance sites. The ATRI-IP will allow facilities to collect, store, process, analyze, visualize, audit, and share scientific, operational, and administrative data and related metadata using standardized platform-independent data models, formats, and taxonomies.
This platform supports multiple NIH sponsored programs including ACTC, ADNI, NIAD, TRC-PAD, ABC-DS, LEADS, TRC-DS.
TRC-PAD Informatics Platform
The Trial-Ready Cohort for Preclinical/Prodromal Alzheimer’s Disease (TRC-PAD) Informatics Platform (TRC-PAD IP) was developed to facilitate the efficient selection, recruitment, and assessment of study participants in support of the TRC-PAD program (Aisen et al., 2020). The TRC-PAD IP provides a secure, scalable, multi-tiered information management platform designed to facilitate high-throughput, cost-effective selection, recruitment, and assessment of TRC-PAD study participants and develop a learning algorithm to select amyloid-bearing participants to participate in trials of early-stage Alzheimer’s disease (Jimenez-Maggiora et al., 2020).
As of 2023, the TRC-PAD IP has supported the enrollment of over 50 thousand participants into the TRC-PAD program. The platform has been extended to support the rapid assessment of novel digital recruitment and retention interventions. Interventions that demonstrate feasibility and effectiveness can be implemented efficiently. The addition of plasma-based biomarker screening is also underway.
ATRI Data Sharing Platform
We are developing a new infrastructure to support open science and data sharing following CAP and FAIR data principles: ATRI DataShare. This infrastructure will serve as the primary repository for making ATRI and ACTC data, tools, and methods available to the scientific community for secondary research. Work on the ATRI DataShare platform has begun with financial and in-kind support from NIH and Gates Ventures. Once complete, ATRI DataShare will:
- Create an informatics infrastructure that allows visualization and linkage of materials to enable precise requests
- Enable selection of data, images, and biospecimens by demographic, genetic, and other data specifications
- Enable sharing of images that are de-identified and defaced (where necessary)
- Package data sets with R code to facilitate analysis and reproducibility
- Share extensive documentation including publications, protocols, consent forms, and data dictionaries